Table of Contents
Weights in Models Fitted with the rma.mv() Function
One of the fundamental concepts underlying a meta-analysis is the idea of weighting: More precise estimates are given more weight in the analysis then less precise estimates. In 'standard' equal- and random-effects models (such as those that can be fitted with the rma() function), the weighting scheme is quite simple and covered in standard textbooks on meta-analysis. However, in more complex models (such as those that can be fitted with the rma.mv() function), the way estimates are weighted is more complex. Here, I will discuss some of those intricacies.
Models Fitted with the rma() Function
To start, let's discuss the standard models first. Here, we are meta-analyzing a set of independent estimates and so we can use the rma() function. For illustration purposes, I will use (once again) the BCG vaccine dataset. We start by computing the log risk ratios and corresponding sampling variances for the 13 trials included in this dataset.
library(metafor) dat <- dat.bcg dat <- escalc(measure="RR", ai=tpos, bi=tneg, ci=cpos, di=cneg, data=dat, slab=paste0(author, ", ", year)) dat
trial author year tpos tneg cpos cneg ablat alloc yi vi 1 1 Aronson 1948 4 119 11 128 44 random -0.8893 0.3256 2 2 Ferguson & Simes 1949 6 300 29 274 55 random -1.5854 0.1946 3 3 Rosenthal et al 1960 3 228 11 209 42 random -1.3481 0.4154 4 4 Hart & Sutherland 1977 62 13536 248 12619 52 random -1.4416 0.0200 5 5 Frimodt-Moller et al 1973 33 5036 47 5761 13 alternate -0.2175 0.0512 6 6 Stein & Aronson 1953 180 1361 372 1079 44 alternate -0.7861 0.0069 7 7 Vandiviere et al 1973 8 2537 10 619 19 random -1.6209 0.2230 8 8 TPT Madras 1980 505 87886 499 87892 13 random 0.0120 0.0040 9 9 Coetzee & Berjak 1968 29 7470 45 7232 27 random -0.4694 0.0564 10 10 Rosenthal et al 1961 17 1699 65 1600 42 systematic -1.3713 0.0730 11 11 Comstock et al 1974 186 50448 141 27197 18 systematic -0.3394 0.0124 12 12 Comstock & Webster 1969 5 2493 3 2338 33 systematic 0.4459 0.5325 13 13 Comstock et al 1976 27 16886 29 17825 33 systematic -0.0173 0.0714
Variable yi contains the log risk ratios and variable vi the corresponding sampling variances.
We now fit equal- and random-effects models to these estimates.
res.ee <- rma(yi, vi, data=dat, method="EE") res.re <- rma(yi, vi, data=dat)
Next, we extract the weights assigned to the estimates in these two models with the weights() function (the values are given in percent) and assemble these values in a matrix with two columns.
w.ee.re <- cbind( paste0(formatC(weights(res.ee), format="f", digits=1, width=4), "%"), paste0(formatC(weights(res.re), format="f", digits=1, width=4), "%"))
Finally, we create a forest plot of the estimates. We add a bit of space at the bottom (by adjusting ylim), add the summary estimates from the two models to the bottom (with the addpoly() function), and also add the weight information from the matrix above as additional elements to the plot (via the ilab and ilab.xpos arguments). Some extra column labels are added with text() and the segments() function is used to add the line under the 'Weights' column label.
forest(dat$yi, dat$vi, xlim=c(-11,5), ylim=c(-3,17), top=4, atransf=exp, at=log(c(1/16, 1/4, 1, 4, 8)), digits=c(2L,4L), ilab=w.ee.re, ilab.lab=c("EE Model","RE Model"), ilab.xpos=c(-6,-4)) abline(h=0) addpoly(res.ee, row=-1) addpoly(res.re, row=-2) text(-5, 16, "Weights", font=2) segments(-7, 15.5, -3, 15.5)
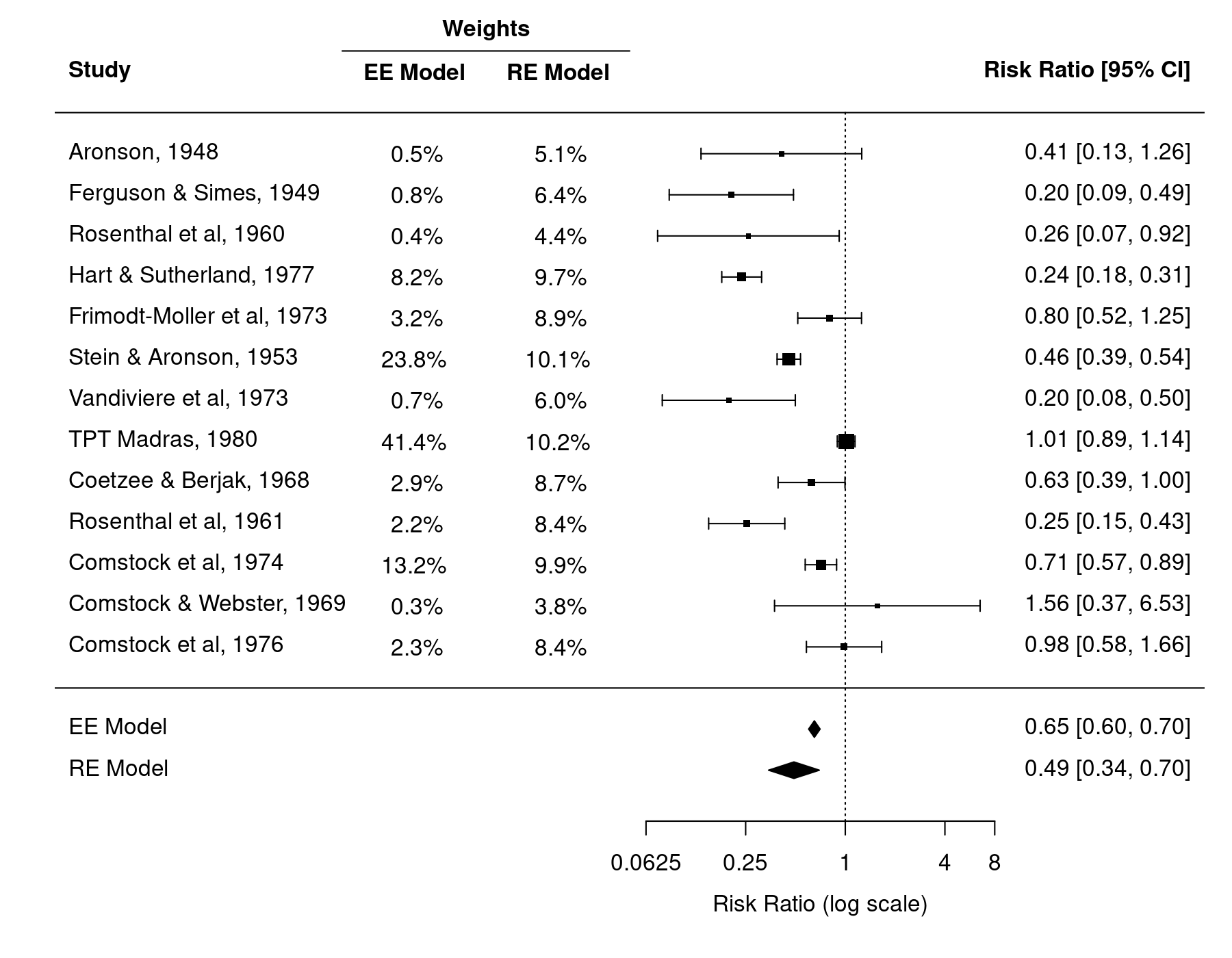
In the equal-effects model, the weights given to the estimates are equal to $w_i = 1 / v_i$, where $v_i$ is the sampling variance of the $i$th study. This is called 'inverse-variance weighting' and can be shown to be the most efficient way of weighting the estimates (i.e., the summary estimate has the lowest possible variance and is therefore most precise). As a result, the estimates with the lowest sampling variances, namely the ones from Stein and Aronson (1953), TPT Madras (1980), and Comstock et al (1974) are given considerably more weight than the rest of the studies.1) Together, these three studies receive almost 80% of the total weight and therefore exert a great deal of influence on the summary estimate. Especially the TPT Madras study 'pulls' the estimate to the right (closer to a risk ratio of 1).
In the RE model, the estimates are weighted with $w_i = 1 / (\hat{\tau}^2 + v_i)$. Therefore, not only the sampling variance, but also the (estimated) amount of heterogeneity (i.e., the variance in the underlying true effects) is taken into consideration when determining the weights. When $\hat{\tau}^2$ is large (relative to the size of the sampling variances), then the weights actually become quite similar to each other. Hence, smaller (less precise) studies may receive almost as much as weight as larger (more precise) studies. We can in fact see this happening in the present example. While the three studies mentioned above still receive the largest weights, their weights are now much more similar to those of the other studies. As a result, the summary estimate is not as strongly pulled to the right by the TPT Madras study.
The weights used in equal- and random-effects models are the inverse of the model-implied variances of the observed outcomes. For example, in the RE model, the model considers two sources of variability that affect the observed outcomes: sampling variability ($v_i$) and heterogeneity ($\hat{\tau}^2$). The sum of these two sources of variability is $\hat{\tau}^2 + v_i$ and the weights are therefore $w_i = 1 / (\hat{\tau}^2 + v_i)$. The summary estimate is then simply the weighted average of the estimates, namely $$\hat{\mu} = \frac{\sum_{i=1}^k w_i y_i}{\sum_{i=1}^k w_i}.$$ By comparing
wi <- 1 / (res.re$tau2 + dat$vi) sum(wi * dat$yi) / sum(wi)
[1] -0.7145323
with the summary estimate from the random-effects model, namely
coef(res.re)
[1] -0.7145323
we can see that this is correct.
Models Fitted with the rma.mv() Function
Models fitted with the rma.mv() function will typically be more complex than those fitted with rma().2) In particular, they will usually involve multiple random effects and possibly correlated sampling errors. See Konstantopoulos (2011) and Berkey et al. (1998) for two illustrative examples. I will use the former example to discuss how the weighting works in such models.
The example involves studies that were conducted at schools that in turn were nested within a higher-ordering grouping variable (districts). This leads to a multilevel structure that we want to account for in the analysis. First, let's copy the data into dat (just to save some typing down the road) and examine the first 8 rows of the dataset. We see that there are 4 studies in the first and second district each.
dat <- dat.konstantopoulos2011 head(dat, 8)
district school study year yi vi 1 11 1 1 1976 -0.180 0.118 2 11 2 2 1976 -0.220 0.118 3 11 3 3 1976 0.230 0.144 4 11 4 4 1976 -0.300 0.144 5 12 1 5 1989 0.130 0.014 6 12 2 6 1989 -0.260 0.014 7 12 3 7 1989 0.190 0.015 8 12 4 8 1989 0.320 0.024
Variable yi contains the standardized mean differences and vi the corresponding sampling variances. To analyze these data, we use a multilevel model with random effects for districts and schools within districts.
res <- rma.mv(yi, vi, random = ~ 1 | district/school, data=dat) res
Multivariate Meta-Analysis Model (k = 56; method: REML)
Variance Components:
estim sqrt nlvls fixed factor
sigma^2.1 0.0651 0.2551 11 no district
sigma^2.2 0.0327 0.1809 56 no district/school
Test for Heterogeneity:
Q(df = 55) = 578.8640, p-val < .0001
Model Results:
estimate se zval pval ci.lb ci.ub
0.1847 0.0846 2.1845 0.0289 0.0190 0.3504 *
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
The model now considers three sources of variability: Between-district heterogeneity ($\hat{\sigma}^2_1$), within-district heterogeneity ($\hat{\sigma}^2_2$), and sampling variability ($v_i$). The model-implied variances of the estimates are then the sum of these three sources of variability (i.e., $\hat{\sigma}^2_1 + \hat{\sigma}^2_2 + v_i$). However, the summary estimate from the model in not simply the weighted average of the estimates with weights given by $w_i = 1 / (\hat{\sigma}^2_1 + \hat{\sigma}^2_2 + v_i)$. This weighted average is:
wi <- 1 / (sum(res$sigma2) + dat$vi) sum(wi * dat$yi) / sum(wi)
[1] 0.127597
But this is not equal to the summary estimate computed by rma.mv():
coef(res)
[1] 0.1847132
The two values are not the same, because the model also implies a certain amount of covariance between the effects, which is also taken into consideration when computing the summary estimate. We can examine the model implied marginal variance-covariance (var-cov) matrix of the estimates (or here, the first 8 rows and columns) with:
round(vcov(res, type="obs")[1:8,1:8], 3)
1 2 3 4 5 6 7 8 1 0.216 0.065 0.065 0.065 0.000 0.000 0.000 0.000 2 0.065 0.216 0.065 0.065 0.000 0.000 0.000 0.000 3 0.065 0.065 0.242 0.065 0.000 0.000 0.000 0.000 4 0.065 0.065 0.065 0.242 0.000 0.000 0.000 0.000 5 0.000 0.000 0.000 0.000 0.112 0.065 0.065 0.065 6 0.000 0.000 0.000 0.000 0.065 0.112 0.065 0.065 7 0.000 0.000 0.000 0.000 0.065 0.065 0.113 0.065 8 0.000 0.000 0.000 0.000 0.065 0.065 0.065 0.122
The diagonal elements of this matrix are indeed equal to $\hat{\sigma}^2_1 + \hat{\sigma}^2_2 + v_i$:
round(sum(res$sigma2) + dat$vi[1:8], 3)
[1] 0.216 0.216 0.242 0.242 0.112 0.112 0.113 0.122
But there are also non-zero off-diagonal elements. In particular, note how the first two districts (with 4 estimates each) result in $4 \times 4$ blocks in this matrix. The off-diagonal elements in these blocks are covariances, which are equal to $\hat{\sigma}^2_1$ (i.e., the estimated between-district variance component) in this type of model. Hence, the first 6 rows and columns of the var-cov matrix are equal to $$M = \begin{bmatrix}
\hat{\sigma}^2_1 + \hat{\sigma}^2_2 + v_1 & \hat{\sigma}^2_1 & \hat{\sigma}^2_1 & \hat{\sigma}^2_1 & & \\
\hat{\sigma}^2_1 & \hat{\sigma}^2_1 + \hat{\sigma}^2_2 + v_2 & \hat{\sigma}^2_1 & \hat{\sigma}^2_1 & & \\
\hat{\sigma}^2_1 & \hat{\sigma}^2_1 & \hat{\sigma}^2_1 + \hat{\sigma}^2_2 + v_3 & \hat{\sigma}^2_1 & & \\
\hat{\sigma}^2_1 & \hat{\sigma}^2_1 & \hat{\sigma}^2_1 & \hat{\sigma}^2_1 + \hat{\sigma}^2_2 + v_4 & & \\
& & & & \hat{\sigma}^2_1 + \hat{\sigma}^2_2 + v_5 & \hat{\sigma}^2_1 \\
& & & & \hat{\sigma}^2_1 & \hat{\sigma}^2_1 + \hat{\sigma}^2_2 + v_6 \\
\end{bmatrix}.$$ As in the documentation of the rma.mv() function (see help(rma.mv)), this matrix is referred to as $M$ (for the Marginal var-cov matrix of the estimates implied by the model).
To compute the summary estimate, we need to take the inverse of this matrix. As a result, we not only have weights, but actually an entire weight matrix. We can inspect this matrix (or the first 8 rows and columns) with:
round(weights(res, type="matrix")[1:8,1:8], 3)
1 2 3 4 5 6 7 8 1 5.533 -1.102 -0.939 -0.939 0.000 0.000 0.000 0.000 2 -1.102 5.533 -0.939 -0.939 0.000 0.000 0.000 0.000 3 -0.939 -0.939 4.857 -0.801 0.000 0.000 0.000 0.000 4 -0.939 -0.939 -0.801 4.857 0.000 0.000 0.000 0.000 5 0.000 0.000 0.000 0.000 16.664 -4.733 -4.633 -3.898 6 0.000 0.000 0.000 0.000 -4.733 16.664 -4.633 -3.898 7 0.000 0.000 0.000 0.000 -4.633 -4.633 16.412 -3.817 8 0.000 0.000 0.000 0.000 -3.898 -3.898 -3.817 14.414
Since the var-cov matrix is 'block-diagonal', each block in the weight matrix is actually the inverse of the corresponding block from the $M$ matrix (you can check this for example for the first district with round(solve(vcov(res, type="obs")[1:4,1:4]), 3)).
Let $W = M^{-1}$ denote the weight matrix. Also, let $X$ denote the model matrix, which contains the values of the variables corresponding to the fixed effects. In the present case, the model just contains an intercept term and no further predictors/moderators and hence the model matrix is a single column of 1s (which can be extracted with model.matrix(res)). Finally, let $y$ denote the (column) vector with the estimates. Then the fixed effects are estimated with $$b = (X'WX)^{-1} X'Wy.$$ This is a so-called generalized least squares (GLS) estimator. For the present model, this yields a single number, namely the estimated intercept (i.e., the pooled/summary estimate). We can check that this works by manually crunching through the matrix algebra.3)
W <- weights(res, type="matrix") X <- model.matrix(res) y <- cbind(dat$yi) solve(t(X) %*% W %*% X) %*% t(X) %*% W %*% y
[1] 0.1847132
This is indeed the same value as we obtained above with coef(res).
For those less familiar with matrix algebra, the equation that provides the summary estimate for such an 'intercept-only model' can also be written in the more familiar summation notation. Let $w_{ij}$ denote the element in the $i$th row and $j$th column of the weight matrix and let $y_i$ denote the $i$th estimate (with $i = 1, \ldots, k$). Then $$\hat{\mu} = \frac{\sum_{i=1}^k (\sum_{j=1}^k w_{ij}) y_i}{\sum_{i=1}^k \sum_{j=1}^k w_{ij}}.$$ The denominator of this fraction is just the sum of all elements in the weight matrix. In the numerator, we sum up the weights in each column of the weight matrix (which is the same as taking the sum in each row, since the weight matrix is symmetric) and multiply this sum by the corresponding estimate. This is done for each estimate and the resulting values are summed up. Let's check again that this is correct:
W <- weights(res, type="matrix") sum(rowSums(W) * dat$yi) / sum(W)
[1] 0.1847132
While rowSums(W) takes the sum of all weight values in each row, most of those values are actually zeros. Hence, for each estimate, we really just need to sum up the values that are part of the block to which an estimate belongs. For example, for the first estimate (i.e., dat$yi[1]), we just need to sum up the first four values in the first row, that is, we would sum up:
W[1,1:4]
1 2 3 4 5.5325581 -1.1015344 -0.9394859 -0.9394859
For the second estimate, we would sum up W[2,1:4]. For the fifth estimate (which belongs to the second district), we would sum up W[5,5:8]. And so on.
If we look at the equation above again, we should note that $\hat{\mu}$ is actually still a weighted average of the estimates, but now the weight given to the $i$th estimate is equal to $\sum_{j=1}^k w_{ij}$. These 'row-sum weights' can be obtained with:
weights(res, type="rowsum")[1:8]
1 2 3 4 5 6 7 8 1.824641 1.824641 1.556215 1.556215 2.430599 2.430599 2.379682 2.002198
They are given as percentages. We can use these weights to compute the weighted average of the estimates, which again yields the summary estimate:
wi <- weights(res, type="rowsum") sum(wi * dat$yi) / sum(wi)
[1] 0.1847132
By examining the weight matrix, we can also build some intuition how the model accounts for multiple estimates coming from the same district. Note that the off-diagonal elements in the blocks of the weight matrix are negative. This results in a certain amount of downweighting of the estimates, so that districts contributing many estimates do not automatically receive tons of weight when computing the summary estimate. To illustrate this, we can compute the sum of the row-sum weights within each district. In addition, the table below shows how many estimates each district contributes.
data.frame(k = c(table(dat$district)), weight = tapply(wi, dat$district, sum))
k weight 11 4 6.761714 12 4 9.243078 18 3 8.536540 27 4 9.506693 56 4 9.502062 58 11 10.328499 71 3 8.943803 86 8 10.320158 91 6 9.883123 108 5 9.203041 644 4 7.771289
While district 58, which contributes 11 estimates, does receive the most weight, this is closely followed by district 86, which contributes 8 estimates. And not much further behind are districts 27 and 56, which 'only' contribute 4 estimates. This should alleviate, at least to some extent, worries that districts contributing many estimates are receiving 'too much weight' in the analysis.
Note that the degree of downweighting depends on the covariance between the estimates, which in the present model is simply the between-district heterogeneity. This variance component is quite large in the illustrative example (almost twice as large as the within-district heterogeneity; see res$sigma2[1] / res$sigma2[2]). In cases where this ratio is much smaller, clusters contributing many estimates will receive proportionally more weight (although the sampling variances also play a role here, so many imprecise estimates may still not receive much weight after all).
Conclusions
The example above shows that the weighting scheme underlying more complex models (that can be fitted with the rma.mv() function) is not as simple as in the 'standard' equal- and random-effects models (that can be fitted with the rma() function). Depending on the random effects included in the model (and the var-cov matrix of the sampling errors), the model may imply a certain degree of covariance between the estimates, which needs to be taken into consideration when estimating the fixed effects (e.g., the pooled/summary estimate) and their corresponding standard errors.
Note: James Pustejovsky has written up a very nice blog post which goes even deeper into this topic.
References
Berkey, C. S., Hoaglin, D. C., Antczak-Bouckoms, A., Mosteller, F., & Colditz, G. A. (1998). Meta-analysis of multiple outcomes by regression with random effects. Statistics in Medicine, 17(22), 2537–2550.
Konstantopoulos, S. (2011). Fixed effects and variance components estimation in three-level meta-analysis. Research Synthesis Methods, 2(1), 61–76.
rma.mv() can be used to fit the same models that can be fitted with rma().rma.mv() function therefore uses a Cholesky decomposition for obtaining the estimates of the fixed effects.